-Search query
-Search result
Showing 1 - 50 of 72 items for (author: didier & t)

EMDB-17819: 
XBB 1.0 RBD bound to P4J15 (Local)
Method: single particle / : Duhoo Y, Lau K
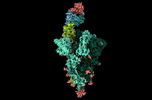
EMDB-17849: 
XBB 1.0 RBD bound to P4J15 (Global)
Method: single particle / : Duhoo Y, Lau K

EMDB-17850: 
SARS-CoV-2 XBB 1.0 closed conformation.
Method: single particle / : Duhoo Y, Lau K

EMDB-17030: 
Helical nucleocapsid of the Respiratory Syncytial Virus
Method: helical / : Gonnin L, Desfosses A, Gutsche I

EMDB-17031: 
Double-ring nucleocapsid of the Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Gutsche I

EMDB-17034: 
Helical nucleocapsid of the N1-370 mutant of the human Respiratory Syncytial Virus
Method: helical / : Gonnin L, Desfosses A, Gutsche I

EMDB-17035: 
Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Eleouet JF, Galloux M, Gutsche I

EMDB-17036: 
Double-headed nucleocapsid of the human Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Gutsche I

EMDB-17037: 
Ring-capped nucleocapsid of the Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Gutsche I

EMDB-17038: 
Stacks of nucleocapsid rings of the N1-370 mutant of the human Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Gutsche I

PDB-8oou: 
Double-ring nucleocapsid of the Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Gutsche I

PDB-8op1: 
Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Eleouet JF, Galloux M, Gutsche I

PDB-8op2: 
Stacks of nucleocapsid rings of the N1-370 mutant of the human Respiratory Syncytial Virus
Method: single particle / : Gonnin L, Desfosses A, Gutsche I

EMDB-14727: 
3D reconstruction of the cylindrical assembly of DnaJA2 by imposing D5 symmetry
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta JM, Muga A

EMDB-14729: 
3D reconstruction of the cylindrical assembly of DnaJA2 without symmetry imposition
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta JM, Muga A

EMDB-14736: 
3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta J, Muga A

PDB-7zhs: 
3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta J, Muga A

EMDB-14706: 
3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F without symmetry imposition
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Muga A, Valpuesta JM

EMDB-16827: 
Condensed RPA-DNA nucleoprotein filament
Method: single particle / : Madru C, Martinez-Carranza M, Sauguet L

PDB-8oel: 
Condensed RPA-DNA nucleoprotein filament
Method: single particle / : Madru C, Martinez-Carranza M, Sauguet L

EMDB-16445: 
RPA tetrameric supercomplex with AROD-OB-1
Method: single particle / : Madru C, Martinez-Carranza M, Legrand P, Sauguet L

EMDB-16826: 
Extended RPA-DNA nucleoprotein filament
Method: single particle / : Madru C, Martinez-Carranza M, Sauguet L

PDB-8c5z: 
RPA tetrameric supercomplex with AROD-OB-1
Method: single particle / : Madru C, Martinez-Carranza M, Legrand P, Sauguet L

PDB-8oej: 
Extended RPA-DNA nucleoprotein filament
Method: single particle / : Madru C, Martinez-Carranza M, Sauguet L

EMDB-16444: 
RPA tetrameric supercomplex from Pyrococcus abyssi
Method: single particle / : Madru C, Martinez-Carranza M, Legrand P, Sauguet L

EMDB-16448: 
Replication Protein A bound to DNA
Method: single particle / : Madru C, Martinez-Carranza M, Legrand P, Sauguet L

PDB-8c5y: 
RPA tetrameric supercomplex from Pyrococcus abyssi
Method: single particle / : Madru C, Martinez-Carranza M, Legrand P, Sauguet L

EMDB-15592: 
BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

PDB-8aqw: 
BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

EMDB-14922: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

PDB-7zrv: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

EMDB-14930: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

EMDB-14947: 
cryo-EM structure of D614 spike in complex with de novo designed binder, full and local maps(addition)
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

PDB-7zsd: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

PDB-7zss: 
cryo-EM structure of D614 spike in complex with de novo designed binder
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

EMDB-15588: 
BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

EMDB-15589: 
Beta SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

EMDB-15590: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

EMDB-15591: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

PDB-8aqs: 
BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

PDB-8aqt: 
Beta SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

PDB-8aqu: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

PDB-8aqv: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)
Method: single particle / : Lau K, Ni D, Beckert B, Nazarov S, Myasnikov A, Pojer F, Stahlberg H, Uchikawa E

EMDB-14141: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

EMDB-14142: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

EMDB-14143: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qti: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qtj: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
